rDNAome - GANLEY Laboratory

Research
Our lab is primarily interested in two kinds of rDNA: ribosomal DNA and random DNA.
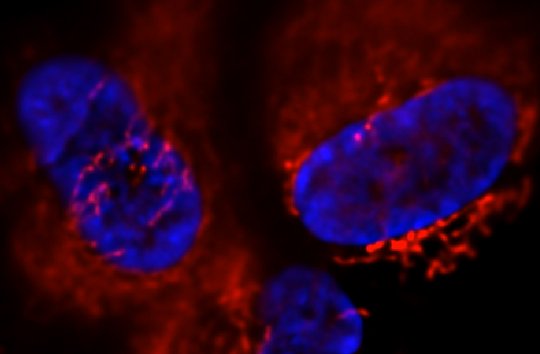
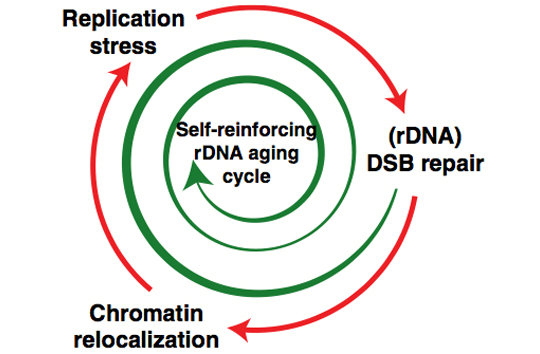
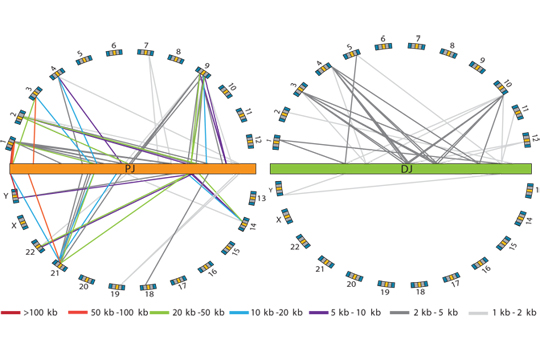
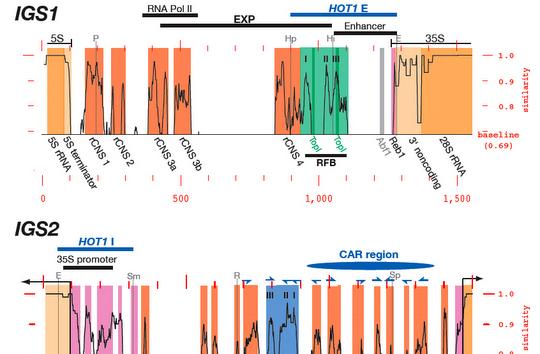
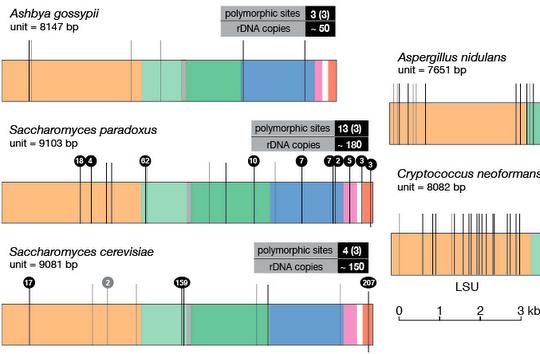
In terms of ribosomal DNA, we are interested in the dynamics and evolution of this fascinating multi-copy locus. The repetitive organization of the ribosomal DNA likely goes back to the last common ancestor of the eukaryotes, thus ribosomal DNA has persisted in this unique and distinctive form for over a billion years, although the reasons why are not clear. Another part of the appeal of the ribosomal DNA repeats is that the repeats within a genome evolve like individuals in a population - they are a population of "individuals" evolving within a population of actual individuals. This gives them a level of dynamism not shared with other genes, and this dynamism is aided-and-abetted by the high level of recombination they undergo, all resulting in them exhibiting an unusual pattern of evolution known as concerted evolution. Encoding most of the RNA that makes up ribosomes, they are obviously essential for life and appear to be under strong transcriptional regulation, for example as stem cells differentiate and as cells become malignant. Despite this, their repetitive nature has made them difficult to study, thus they remain one of the most poorly-studied parts of eukaryote genomes. We are investigating the rDNA using both molecular and computational approaches.
In terms of random DNA, we are interested in synthesizing random DNA to better understand the nature of large eukaryote genomes. In genomes such as our own, only a tiny fraction encodes genes. What does the rest do? When answering this question, people tend to fall into two, opposing camps. One group thinks that most of the rest is useless, and unused, junk DNA. The other thinks most of it is functional - we just haven't figured out what the functions are. The latter viewpoint got a massive boost by observations that genomes tend to be pervasively transcribed - surely this transcription is there for a reason (i.e. making functional, noncoding RNA). However, it was pointed out that this could just be background noise and thus pervasive transcription (and other genomic activities) might not be telling us much. A way to distinguish between these views, as first pointed out by Sean Eddy, is to see what happens if you introduce random DNA into a genome. Thus, we are enzymatically synthesizing random for this very reason. We will also using computational approaches to see whether we can distinguish different parts of genomes based on what genomic activities they exhibit.
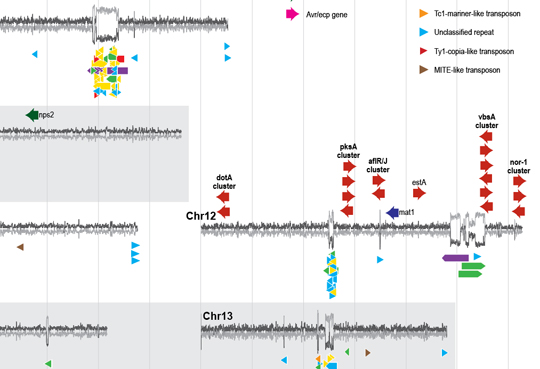
We also maintain interests in repeat biology in general, fungal evolution, yeast fermentation, and mushrooms.
more....
People
 Austen Ganley (Lab head)
Austen Ganley (Lab head)
Austen completed his BSc, BSc(Hons) and PhD in Genetics all at Massey University. From there he voyaged to the USA, to post-doc in the mushroom lab of Rytas Vilgalys at Duke University in North Carolina. After a series of cultural exchanges involving mushroom
more....
News
- Article published in MBE!
We had our article on self-splicing intron dynamics in the fungal genus Epichloë published in Molecular Biology and Evolution! This was based on Jennie's Honours thesis work. The article can be accessed here. (02/04/2025) - News article on research funding
I just had a news article on the recent research funding changes in New Zealand published in Newsroom. (09/02/2025) - Silvio Bonni, new PhD student in synthetic biology, has started!
Welcome to Silvio, who is starting a PhD in synthetic biology looking at selective advantage of reproductive divisions of labour (such as germ-soma separation) in conjunction with Nobuto Takeuchi and Ant Poole. (09/02/2025) - Congratulations, Danni!
Congratulations to Danni on the birth of her second child! (15/01/2025) - Sylvie won SBS Professional Staff Award
Sylvie (together with Carol Wang) won the 2024 SBS Professional Staff Award!! Congratulations to Sylvie and Carol - very well deserved!! (27/11/2024)